|
The Fromme lab is committed to fostering diversity in our lab, in our academic community, and beyond. Science works better when many different perspectives are considered, and no group has a monopoly on the truth. We recognize there is a problem with a striking lack of diversity in academia, and to actively address this problem Fromme lab members lead and participate in several outreach and ambassador programs. Members of the Fromme lab helped to found and lead the MBG Diversity Council, and Fromme lab members have played leadership roles in our Ph.D. Fields’ Graduate Student Associations. |
Recent Fromme Lab News:
|
Research Focus:
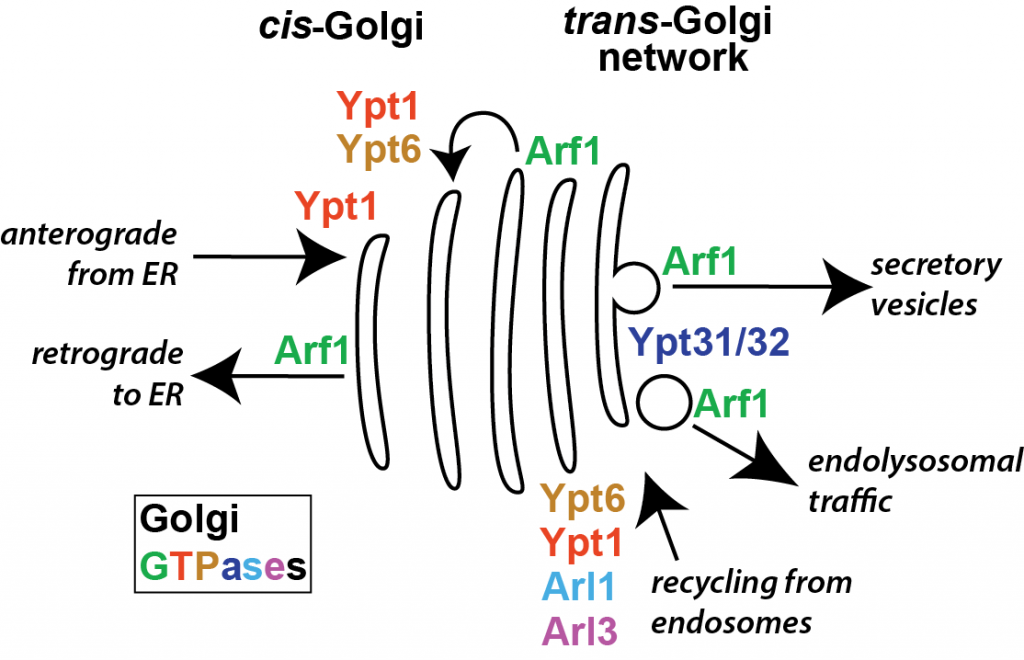
The Golgi complex is the “Grand Central Station” within our cells, serving as the primary sorting organelle at the nexus of the secretory and endocytic trafficking pathways. For example, virtually all proteins that eukaryotic cells display on their surface at the plasma membrane are first synthesized at the endoplasmic reticulum and then trafficked to the Golgi complex. Once at the Golgi, proteins find themselves at a crossroads: they may be trafficked to the plasma membrane, to endocytic organelles, to lysosomal organelles, back to the endoplasmic reticulum, or they may remain within the Golgi complex. We view the Golgi as an excellent model for investigating how decisions are made at the level of an organelle: How does the Golgi maintain homeostasis in the face of constant flux? How does the Golgi respond to changes in cargo load? Does the Golgi communicate with other organelles?
The regulators for all incoming and outgoing Golgi traffic are GTPases of the Rab and Arf families. Our lab has discovered that GTPase activation is regulated at the Golgi through autoinhibition, positive feedback, and GTPase crosstalk mechanisms. Our findings lead to a model for regulation of the Golgi in which multiple GTPases pathways, previously considered to act in isolation, are intimately connected. By investigating how the GTPases and their effectors are regulated using biochemical, structural, and cell biological approaches, we aim to uncover the molecular logic governing regulation of the Golgi at a mechanistic level.
Golgi trafficking is controlled by a discrete set of GTPases